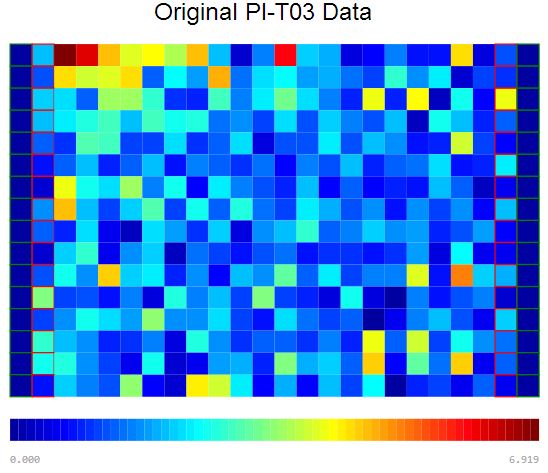
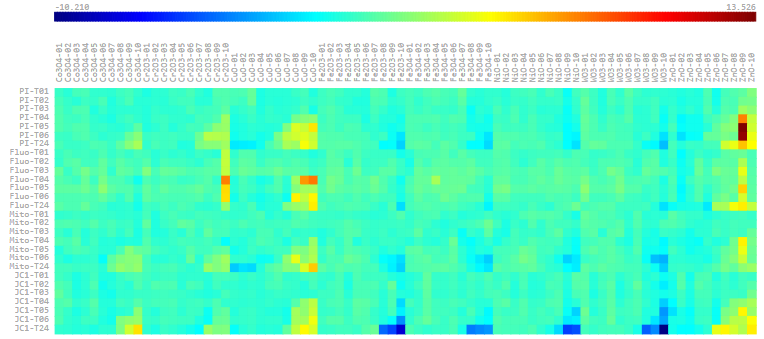
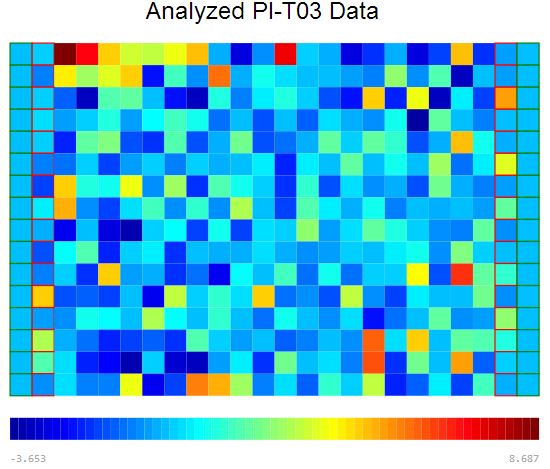
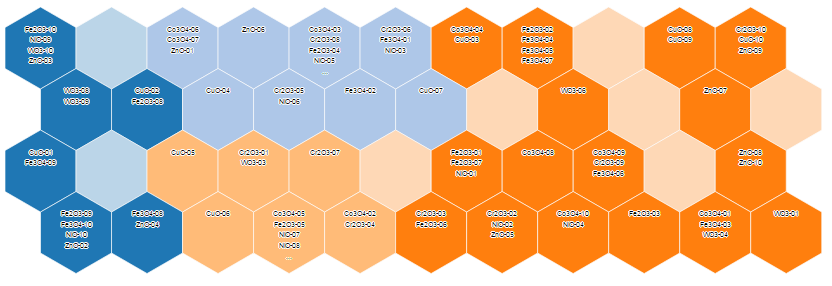
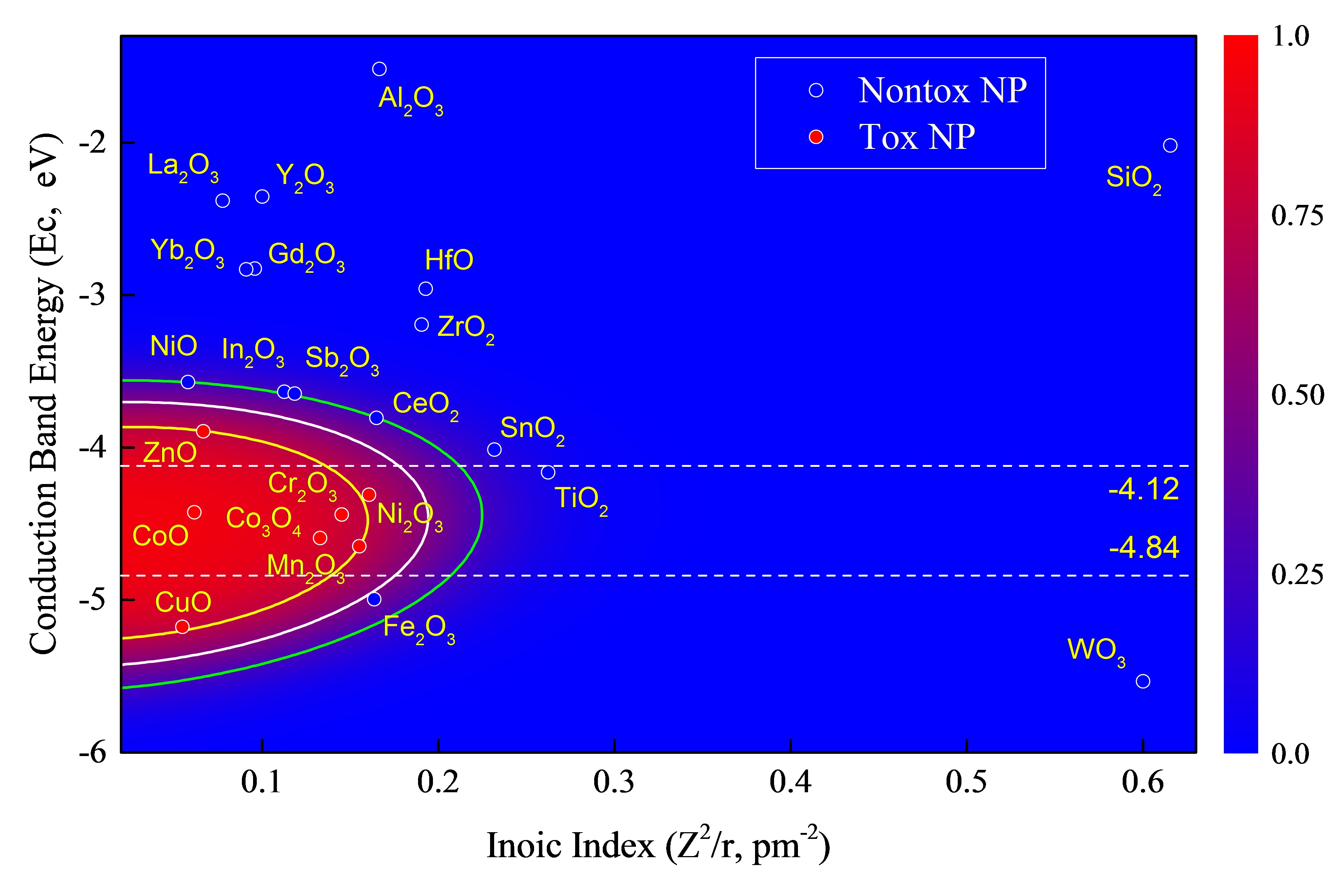
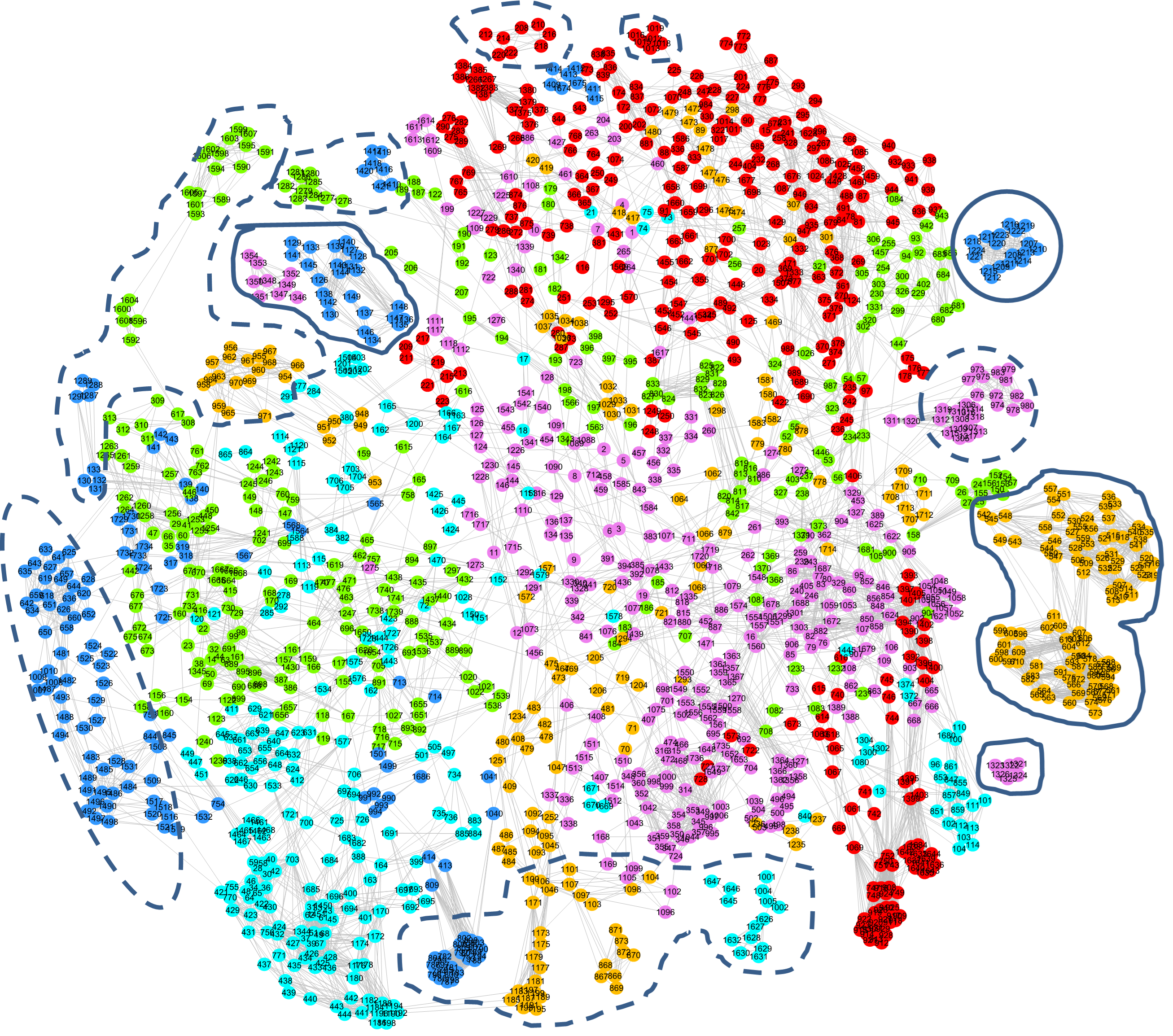
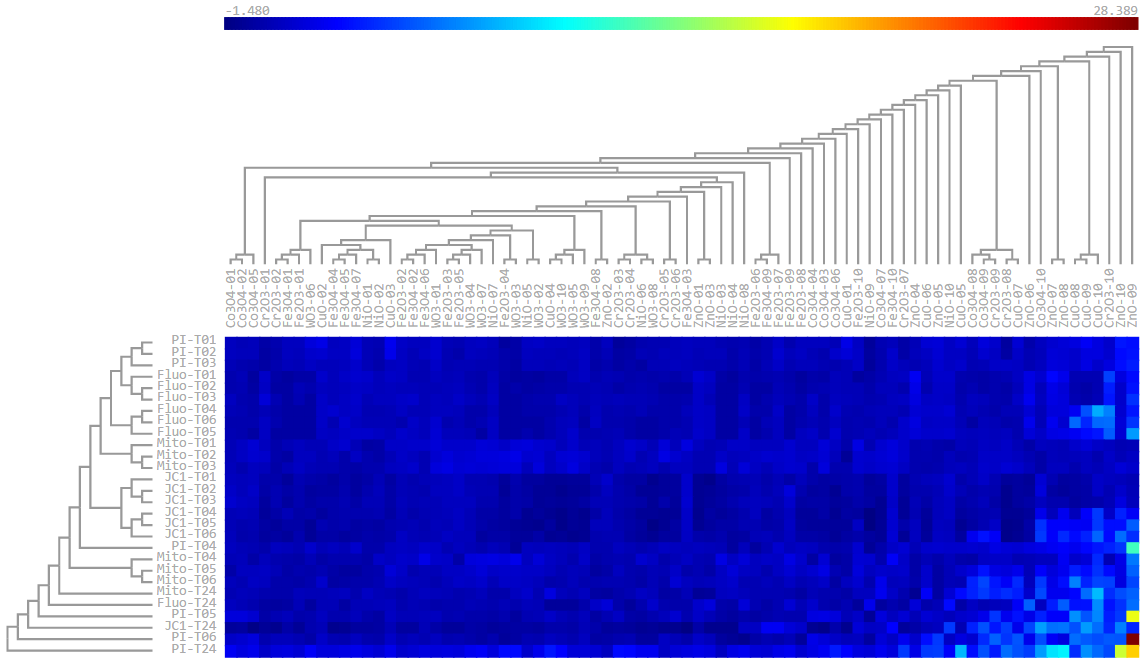
Raw Data Visualization
Plate visualization allows initial visual inspection of HTS and HC datasets to evaluate the consistency of sample replicates and the effectiveness of positive/negative controls. Rapid data visualization can then guide the selection of suitable statistical methods for subsequent data analysis.